HIV-1 reverse transcriptase (RT) is a multifunctional enzyme responsible for the transcription of the RNA genome of the HIV virus into DNA suitable for incorporation within the DNA of human host cells.
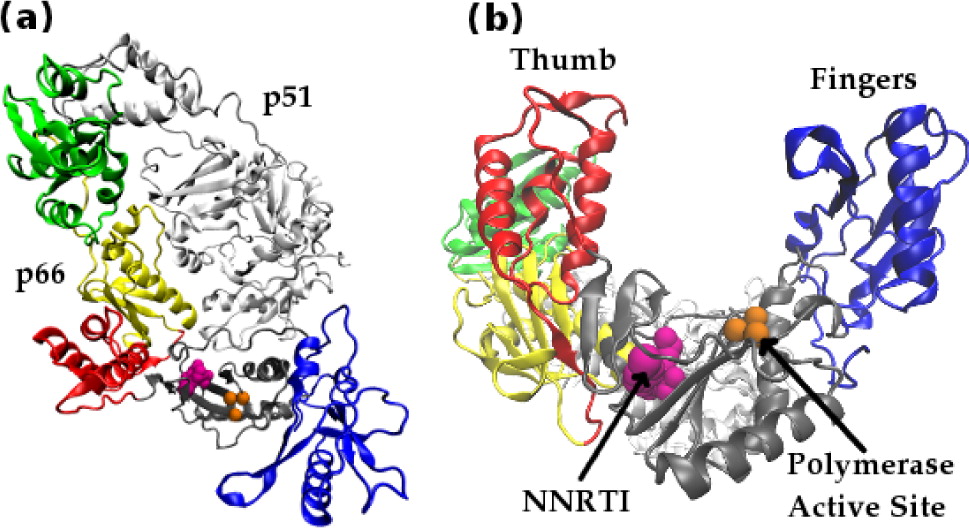
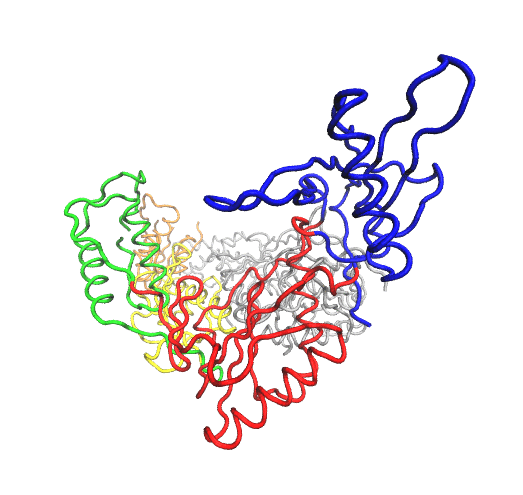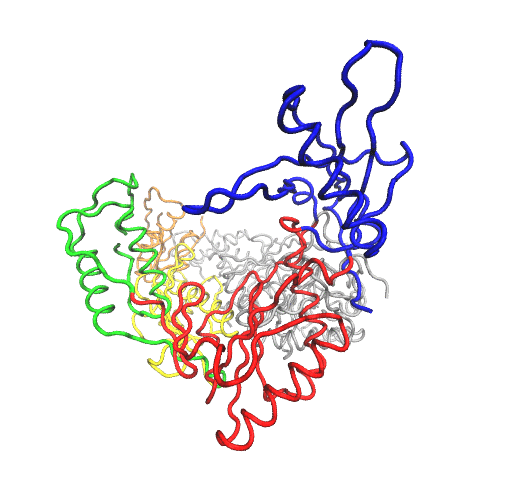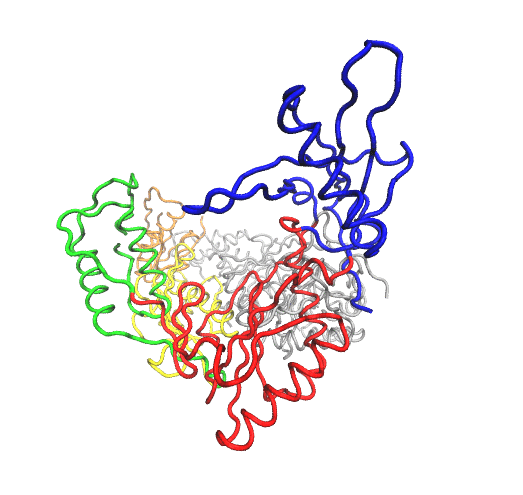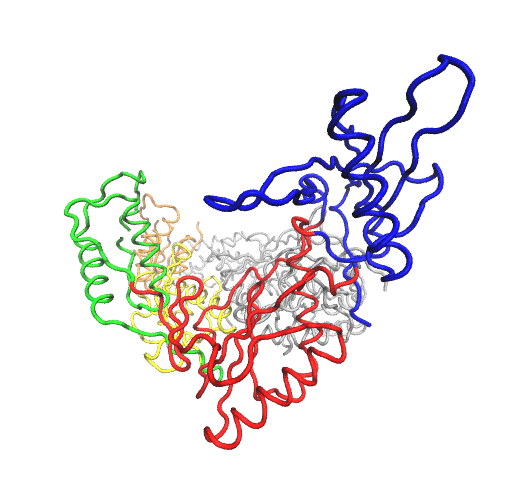
HIV-1 RT is a heterodimer consisting of a p66 and a p51 subunit. In Figure 1 the domains of p66 are individually colored: the fingers are shown in blue, the palm gray, the thumb red, the connection yellow, and the RNaseH green. The p51 subunit is shown in white. The location of the polymerase active site triad is depicted in orange. The location of the bound drug efavirenz (an NNRTI, see below) is shown in magenta. The view along the binding cleft illustrates the resemblance of the p66 subunit to an opened hand.”
The enzyme uses the single stranded RNA viral genome as a template to create a single strand of DNA which is in turn used as a template to create a double stranded DNA (dsDNA) copy of the genome. The dsDNA copy is then ready to be integrated into the chromosomes of host cells. HIV-1 RT has two distinct active sites, known as the polymerase and RNaseH active sites respectively. At the polymerase active site incoming nucleotides matching the template RNA or DNA are incorporated into the growing complementary DNA chain. The RNaseH active site catalyses the breakdown of the RNA genome, freeing the DNA copy to act as a template for the creation of the final double stranded DNA genome. Inhibition of either enzymatic activity compromises viral replication and its crucial role in the HIV life cycle has made it one of the principal targets for antiretroviral drug therapy. Two classes of inhibitor are currently in clinical use, called nucleotide analogue RT inhibitors (NRTIs) and non-nucleoside analogue RT inhibitors (NNRTIs). The former mimic the natural deoxynucleotide triphosphate (dNTP) substrate, binding to nascent DNA chains and acting as chain terminators preventing further elongation. NNRTIs operate allosterically, binding approximately 10 Å from the polymerase active site in a pocket which does not exist in the apo enzyme, inhibiting the enzyme allosterically. I have used molecular dynamics and coarse grained network models to investigate the impact of NNRTIs on the structure and dynamics of HIV-1 RT.
Thumbs Down: Domain Rearrangements in NNRTI Bound RT
It is widely believed that NNRTIs function as “molecular wedges”, disrupting the region between thumb and palm subdomains of the p66 subunit and locking the thumb in a wide-open conformation. Crystal structure data suggest that the binding of NNRTIs forces RT into a wide-open conformation in which the separation between the thumb and fingers subdomains is much higher than in the apo structure. Using ensemble molecular dynamics simulations (aggregate sampling 600 ns), we have captured RT bound to the NNRTI efavirenz (efz) in a closed conformation similar to that of the apo enzyme, suggesting the constraint of thumb motion is not as complete as previously believed. Our simulations confirm that a conformational distribution across open and closed states must exist in the drug-bound enzyme and that allosteric modulation is effected via the alteration of the kinetic landscape of conformational transitions upon drug-binding.
- D. W. Wright, S. K. Sadiq, G. De Fabritiis and P. V. Coveney, “Thumbs down for HIV: Domain level rearrangements do occur in the NNRTI bound HIV-1 Reverse Transcriptase”, Journal of the American Chemical Society, 2012, 134 (31), DOI:10.1021/ja301565k
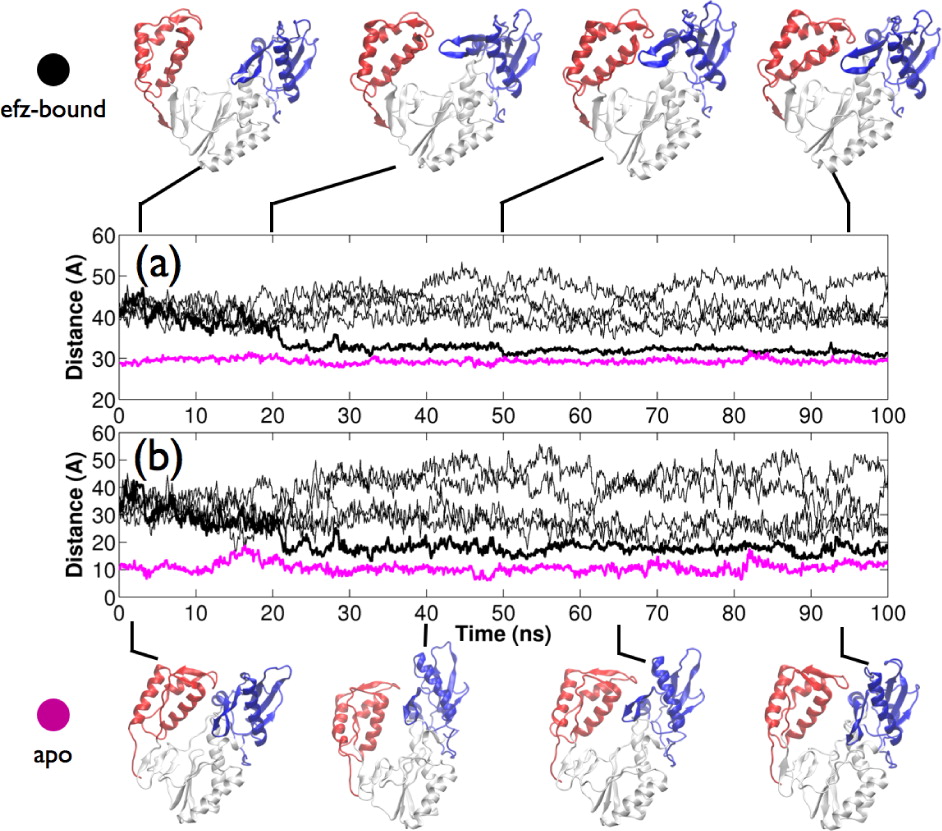
Global Dynamics of NNRTI bound HIV-1 RT Bound
Using both coarse grained network models and atomistic molecular dynamics we explored changes in protein dynamics induced by NNRTI binding. We identify changes in the flexibility and conformation of residue Glu396 in the RNaseH primer grip which could provide an explanation for the acceleration in RNaseH cleavage rate observed experimentally in NNRTI bound HIV-1 RT. Unlike previous network models we did not see significant differences in the motions predicted by the network models for different NNRTIs, rather we see that the motions are altered by differences in the conformation captured crystallographically. Additionally, we suggest a plausible path for conformational and dynamic changes to be communicated from the vicinity of the NNRTI binding pocket to the RNaseH at the other end of the enzyme.
- D. W. Wright, B. A. Hall, P. Kellam and P. V. Coveney, “Global Conformational Dynamics of HIV-1 Reverse Transcriptase Bound to Non Nucleoside Inhibitors”, Biology, 2012, 1 (2), DOI: 10.3390/biology1020222